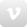Michel Valim
One of the more poorly understood aspects of avian ecology is the role that diseases play in avian life histories. In recent years our lack of understanding of the subject has come to the forefront because of avian flu (H5N1) outbreaks that started in east Asia and soon spread around the world, accompanied by the transmission of the mutated flu virus from birds to humans. Since that time, the study of avian disease ecology has increased tremendously. Here at the Field Museum, our expeditions now include sampling that allows us to study a variety of parasites and pathogens across a large sampling of bird (and mammal) species, in part for the Emerging Pathogens Project.
One set of pathogens that we have been actively studying is blood parasites. There are many types of avian blood parasites, including malarial parasites (Plasmodium, Haemoproteus, and Parahaemoproteus), Leucocytozoon, Trypanosomes, and others. These infections can be detected using two complementary methods, and our field sampling protocols provide the materials needed for both methods. One method could be called the old-fashioned way. A drop of blood is smeared on a microscope slide and then is stained, and a scientist looks under a microscope to detect the infection visually in the cells. The other way is to detect the infections genetically, by running PCR-based DNA screening, which we do in the Field Museum’s Pritzker Lab for Molecular Systematics and Evolution. Not surprisingly, the best way to ensure accuracy is to use both methods.
A remarkable and consistent finding in avian malaria research is just how widespread some strains are. Some strains of these blood parasites are found in a wide variety of species and all over the world, whereas others are extremely specific to one lineage of hosts (for example, one type of Haemoproteus is found only in pigeons and doves).
We are also studying various groups of ectoparasites, including ectoparasitic chewing lice. Like the blood parasites mentioned above, these ectoparasites have lots of variablity in host specificity (how many hosts a parasite is able to infect). Some chewing lice species infect only a single host species, whereas others are capable of infecting many different host species. One of the goals of our research on this interesting group of parasites is to understand the ecological correlates of host specificity. In other words, we are trying to understand what factors allow some parasites to infect only a single host whereas others can infect many hosts. Another question we are interested in asking is what ecological factors affect a parasite's ability to switch hosts.
To answer these an other questions about avian parasite and pathogen evolutionary history we typically use DNA sequences to reconstruct evolutionary histories (evolutionary trees) of the parasites and their avian hosts and then use these to make inferences about how the hosts and their parasites have evolved over evolutionary time. Using these evolutionary reconstructions helps us to further understand evolutionary phenomena such as host-switching and host-specificity, which is important for a broader understanding of parasite and pathogen evolution and ecology.

These are two genera of lice collected from a Willet (Tringa semipalmata) that was found dead on the Chicago lakefront. Looking closely, you can see what they've been eatings: blood (left) and feather barbs (right). Photo by Jason Weckstein.